MAPEO GENÉTICO
MA 102 VARIEDADES ARGENTINAS
Esta pequeña población de mapeo por asociación se desarrolló originalmente para el estudio de caracteres asociados a la adaptación del cultivo y a componentes de rendimiento, los resultados mostrados en esta página son los obtenidos y publicados por Basile et al. (2019).
Información de la colección:
Un conjunto de 102 variedades de trigo pan descritos anteriormente (Vanzetti et al., 2013) que incluye variedades antiguas (como el cv. Barletta 77, en 1927) y cultivares comerciales recientes (hasta 2010) fueron seleccionados entre las principales empresas de mejoramiento de trigo en Argentina y utilizado para la construcción de bloques de haplotipos y análisis GWAS. Las semillas fueron obtenidas del Banco de Germoplasma de trigo de la Estación Experimental INTA Marcos Juárez.
Tipo de Población: Mapeo por Asociación.
Tamaño de la Población: 102 genotipos
Origen: EEA INTA Marcos Juarez. Responsable del proyecto Dr. Leonardo Vanzetti
Plataforma de genotipado: Axiom® 35K SNP Wheat Breeder’s Array (Affimetrix)
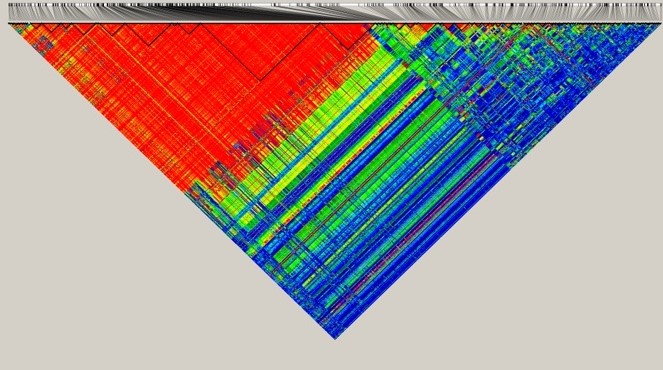
Esquema del cromosoma 1B donde se observa el alto DL provocado por la presencia de la translocación de centeno 1BL/1RS en alta frecuencia en el germoplasma argentino de trigo pan utilizado.
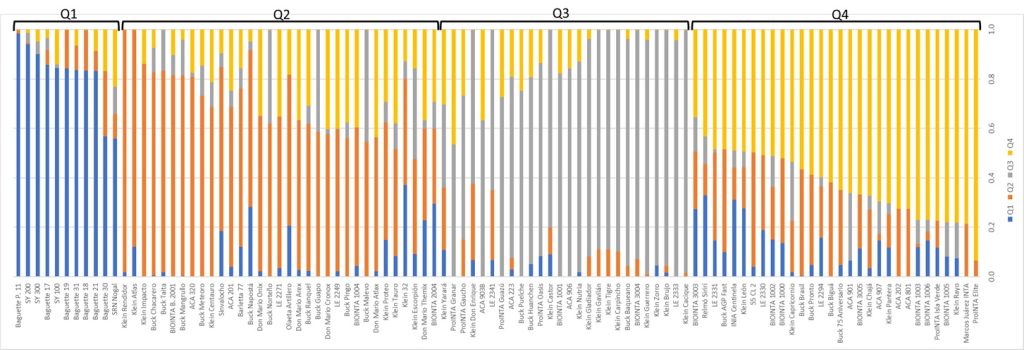
Figura 1: Análisis de la estructura de la colección de 102 cultivares de trigo hexaploides argentinos. El análisis de estructura mostró cuatro subpoblaciones hipotéticas representadas por diferentes colores.
Estructura de la población (Matriz Q)
la estructura genética en el nivel de población utilizando el software R STRUCTURE (François, 2016)
Análisis de haplotipos
La estructura del haplotipo basada en los marcadores SNP para cada cromosoma de trigo se evaluó utilizando el software Haploview 4.2. (Barrett et al., 2004). El software define bloques de haplotipos (HB) y proporciona el número de haplotipos y su longitud física (pb) para cada bloque. Los HB se convirtieron en una tabla usando un script de Python propio disponible en GitHub.
Análisis de asociación
Los análisis de GWAS se realizaron utilizando 4.516 haplotipos (provenientes de 1.268 HB) y 495 marcadores genéticos / SNP informativos anclados al genoma del trigo. Para los análisis se utilizó el método SUPER (Wang et al., 2014) implementados en el paquete R GAPIT (Lipka et al., 2012). Para reducir asociaciones espurias se utilizó en el modelo la estructura genética en la población (valores Q). A partir del GWAS para cada carácter, se seleccionó haplotipos / marcadores que fueron significativos (P < 0,05) en cuatro ambientes, con al menos un ambiente con diferencias altamente significativas (P < 0,001). Todos los haplotipos / marcadores que satisfacen estos criterios son presentados en la tabla que esta debajo.
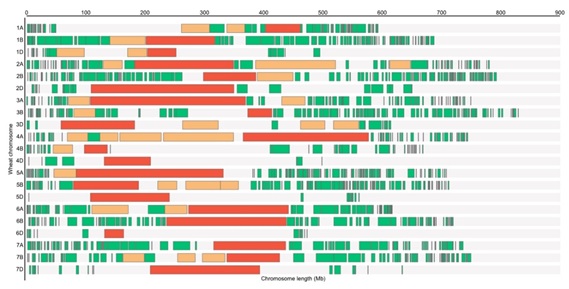
Figura 2: Tamaños y posición de los bloques de haplotipos (HB) en cada cromosoma para la colección de trigo hexaploide argentino. Los HB rojos indican un tamaño > 30 Mb, los tamaños de HB naranjas entre 10 y 30 Mb y los tamaños de HB verdes <10 Mb.
Basado en la fórmula descrita en (Zhang et al., 2018), la probabilidad de que un haplotipo / marcador sea estadísticamente significativo por azar, simultáneamente para cuatro GWAS fue estimado en menos de 1,25E-07 (0,05 x 0,05 x 0,05 x 0,001). Basado en este número, la probabilidad de al menos un error en los 5.011 haplotipos / SNP se estimó como 1 – (1 -1,25E-07) 5.011 = 0,00063 (por carácter evaluado).
Caracteres fenotipados en la población hasta el momento y sobre los cuales se realizó la búsqueda de QTL
DE: Días de siembra a espigazón.
AP: Altura de planta.
FEm = Fertilidad de Espiga a madurez (granos/ gr chaff).
NGE: Número de granos por espiga (número).
PMG: Peso de mil Granos (g).
Resultados del análisis GWAS de la población
| Carácter | Haplotipo/ SNP | Cromosoma | Inicio (Mb) | Fin (Mb) | Frecuencia (%) | Efecto sobre carácter | Alelo | SNP | Azul 2013 | Azul 2014 | Balcarce 2013 | Balcarce 2014 | Balcarce 2015 | Marcos Juárez 2011 | Marcos Juárez 2012 | Marcos Juárez 2013 | Marcos Juárez 2014 | Marcos Juárez 2015 | Marcos Juárez 2016 | Colocalización | Referencia |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FEm | B32-Hap4 | 1A | 482,3 | 487 | 6 | Aumenta | CCTTCTTAC | AX94938408, AX95024082, AX94644800, AX94507352, AX95120675, AX94448644, AX94997258, AX95629264, AX94757176 | * | ns | * | * | ** | - | ns | ns | ns | ns | ns | _ | Basile et al., 2019 |
| FEm | B19-Hap3 | 2A | 70,2 | 72,9 | 12 | Aumenta | GTCG | AX94639168, AX95096480, AX94381449, AX94400797 | * | ns | * | * | ns | - | ** | ns | * | ns | ns | _ | Basile et al., 2019 |
| FEm | B23-Hap1 | 2A | 83,8 | 84,5 | 52 | Disminuye | AT | AX95631188, AX94466696 | * | * | * | * | ns | - | * | ns | ns | * | * | _ | Basile et al., 2019 |
| FEm | B26-Hap3 | 2A | 102 | 102,7 | 36 | Aumenta | AAG | AX94880269, AX95164703, AX94669822 | * | ** | ** | * | ns | - | ns | ns | ns | ns | * | _ | Basile et al., 2019 |
| FEm | B30-Hap4 | 2A | 165,8 | 182,1 | 7 | Aumenta | TTCCGTCC | AX94774467, AX94576255, AX94849076, AX94692672, AX95115244, AX94448613, AX94724618, AX94577367 | ** | ns | * | * | ns | - | ns | * | ns | ns | ns | _ | Basile et al., 2019 |
| FEm | B49-Hap2 | 2A | 704,8 | 705,8 | 15 | Aumenta | GC | AX94720460, AX94399675 | * | * | * | ** | * | - | ns | ns | ns | ns | ns | NKS | Basile et al., 2019 |
| FEm | AX94874921 | 2A | 707,1 | - | 16 | Aumenta | A | AX94874921 | ns | ns | ns | ns | ns | - | ** | ns | * | * | * | _ | Basile et al., 2019 |
| FEm | B55-Hap1 | 2A | 716,8 | 718,9 | 12 | Aumenta | CAATC | AX95096362, AX94982819, AX94661746, AX94535413, AX94433820 | * | *** | ** | * | * | - | ns | ns | ns | ns | ns | _ | Basile et al., 2019 |
| FEm | B14-Hap4 | 3B | 23 | 24 | 6 | Aumenta | GGTC | AX94624219, AX95189982, AX94413646, AX94873710 | ns | ns | ** | * | * | - | ** | ns | ns | * | ns | _ | Basile et al., 2019 |
| FEm | AX94869767 | 4A | 606,4 | - | 18 | Aumenta | C | AX94869767 | * | ns | * | * | ns | - | * | * | ns | * | * | _ | Basile et al., 2019 |
| FEm | B35-Hap3 | 4A | 625,7 | 625,9 | 23 | Aumenta | GT | AX94952227, AX94629424 | ** | * | ** | * | * | - | ns | ns | ns | ns | ns | _ | Basile et al., 2019 |
| FEm | B33-Hap4 | 5A | 445,2 | 445,2 | 7 | Aumenta | GC | AX95632187, AX94682497 | ** | * | * | * | ns | - | ns | ns | ns | ns | ns | PH | Basile et al., 2019 |
| FEm | B43-Hap1 | 5A | 476,4 | 476,7 | 15 | Aumenta | GACCGCG | AX95132574, AX94876698, AX94638131, AX94983398, AX94955528, AX94430148, AX95114243 | * | ns | * | * | ns | - | ns | ** | ns | * | ns | _ | Basile et al., 2019 |
| FEm | B24-Hap2 | 6A | 205,1 | 233,3 | 17 | Aumenta | TCAA | AX95141224, AX94593271, AX94493694, AX95652741 | *** | * | * | ns | ns | - | ns | ns | ns | ns | * | TKW | Basile et al., 2019 |
| FEm | B28-Hap4 | 6A | 448,7 | 454,6 | 9 | Disminuye | GATC | AX95074798, AX94441458, AX95076677, AX94912807 | * | * | * | ** | * | - | * | ns | ns | ns | ns | _ | Basile et al., 2019 |
| FEm | B29-Hap2 | 6A | 455,6 | 467 | 25 | Aumenta | GATC | AX95074798, AX94441458, AX95076677, AX94912807 | *** | ns | * | * | ns | - | ns | ns | * | ns | * | _ | Basile et al., 2019 |
| FEm | B36-Hap2 | 7A | 119 | 126 | 10 | Aumenta | CCGG | AX94410775, AX94436598, AX94578662, AX94404264 | * | ns | * | ** | * | - | ns | ns | ns | ns | * | _ | Basile et al., 2019 |
| PMG | B5-Hap3 | 2D | 17,6 | 18,2 | 7 | Disminuye | TGA | AX94672889, AX94814880, AX95098966 | * | * | ns | ns | ns | - | ns | ns | * | * | ** | _ | Basile et al., 2019 |
| PMG | B12-Hap3 | 3A | 46,7 | 52,6 | 5 | Disminuye | CCGACACTCCTA | AX94494425, AX94947888, AX94946504, AX94660985, AX95252995, AX94429024, AX94567917, AX94954991, AX94994948, AX94713555, AX94821666, AX94718027 | ns | * | ns | * | * | - | ** | * | * | ns | ns | _ | Basile et al., 2019 |
| PMG | AX95257035 | 3A | 686,8 | - | 11 | Aumenta | T | AX95257035 | ns | ns | ns | ns | ns | - | * | ns | *** | ** | * | _ | Basile et al., 2019 |
| PMG | B111-Hap4 | 3B | 817,4 | 817,8 | 11 | Aumenta | CCCC | AX94758693, AX94799334, AX94401378, AX95131891 | ns | ns | ns | * | ns | - | ** | ns | * | * | ns | PH | Basile et al., 2019 |
| PMG | AX94459169 | 6A | 0,6 | - | 30 | Aumenta | C | AX94459169 | ns | ** | * | ns | ns | - | ns | * | *** | *** | ** | _ | Basile et al., 2019 |
| PMG | B20-Hap3 | 6A | 63,8 | 74 | 21 | Disminuye | TTCAGCGAGGGGTCTCGGA | AX94684858, AX94600197, AX95008689, AX95012251, AX94815030, AX94396601, AX94722330, AX94840852, AX95142812, AX95025823, AX95179502, AX94422265, AX94494618, AX94790181, AX94449290, AX94774196, AX95258743, AX95022356, AX95631090 | ns | ns | * | ns | ns | - | ns | ns | * | ** | * | _ | Basile et al., 2019 |
| PMG | B21-Hap3 | 6A | 74,5 | 101,3 | 20 | Disminuye | TCATTTTCGTTGC | AX94664414, AX94961629, AX94640976, AX94395941, AX95246731, AX94683058, AX94977405, AX94942752, AX95256931, AX94490150, AX94795446, AX95145282, AX95012377 | ns | ns | * | ns | ns | - | ns | ns | * | ** | * | _ | Basile et al., 2019 |
| PMG | B24-Hap2 | 6A | 205,1 | 233,3 | 17 | Disminuye | TCAA | AX95141224, AX94593271, AX94493694, AX95652741 | ns | ns | ** | ns | ns | - | * | ns | * | *** | * | FE | Basile et al., 2019 |
| PMG | B38-Hap3 | 7B | 626,1 | 627,6 | 5 | Aumenta | TG-TC | AX94754235, AX94843008, AX95171526, AX94419535, AX94746769 | ns | *** | ** | ns | ns | - | ns | * | * | ns | ** | _ | Basile et al., 2019 |
| PMG | B60-Hap2 | 7B | 702,3 | 703,7 | 27 | Aumenta | TT | AX94498540, AX94700055 | ns | ns | ns | * | ns | - | * | * | ** | * | * | _ | Basile et al., 2019 |
| PMG | B60-Hap3 | 7B | 702,3 | 703,7 | 15 | Aumenta | CT | AX94498540, AX94700055 | ns | ns | * | ns | ns | - | * | * | ** | ** | ** | _ | Basile et al., 2019 |
| NGE | B49-Hap2 | 2A | 704,8 | 705,8 | 15 | Aumenta | GC | AX94720460, AX94399675 | * | ns | * | ** | ** | - | ns | ns | * | ns | ns | FE | Basile et al., 2019 |
| NGE | B19-Hap5 | 4A | 231 | 349,3 | 13 | Aumenta | CACAGT | AX94392668, AX94728695, AX94606134, AX94394855, AX94661891, AX94482222 | * | ** | ns | ns | ns | - | ** | * | * | ns | * | _ | Basile et al., 2019 |
| DE | B76-Hap2 | 1B | 643,1 | 645,7 | 35 | Aumenta | TCT | AX94439319, AX94766122, AX95017951 | * | ** | ns | ns | ns | ** | * | * | ns | * | * | _ | Basile et al., 2019 |
| DE | B84-Hap3 | 1B | 667,2 | 667,8 | 17 | Disminuye | CGT | AX95199577, AX94689744, AX94803292 | ns | ns | ** | ** | * | ns | * | * | * | ns | ns | _ | Basile et al., 2019 |
| DE | B89-Hap4 | 1B | 676,8 | 678 | 6 | Disminuye | -GG | AX94431388, AX94537816, AX94546627 | ns | ns | * | ** | * | * | * | ns | * | * | ns | _ | Basile et al., 2019 |
| DE | B40-Hap4 | 2B | 162,9 | 166,9 | 10 | Aumenta | TTTTGTATA | AX94622328, AX94450544, AX94655193, AX94755547, AX95213611, AX95081080, AX94499577, AX95007105, AX94589917 | * | ** | ns | ns | ns | ns | * | ns | * | * | ns | _ | Basile et al., 2019 |
| DE | B62-Hap1 | 2B | 564,2 | 569 | 10 | Aumenta | ACAAAA | AX95146301, AX94459397, AX95685439, AX94967288, AX95235626, AX94770158 | * | ** | ns | ns | ns | * | ns | ns | ns | * | ns | _ | Basile et al., 2019 |
| DE | Ppd-D1 sensitive | 2D | 34 | - | 42 | Aumenta | sensitive | Ppd-D1 | * | * | * | * | * | ** | *** | ** | *** | *** | ** | _ | Basile et al., 2019 |
| DE | B31-Hap1 | 3A | 504,8 | 507,5 | 59 | Disminuye | AGG | AX94818694, AX94989495, AX94664169 | ns | ns | * | ns | * | ** | * | * | * | * | * | _ | Basile et al., 2019 |
| DE | B49-Hap3 | 3A | 631,2 | 633,1 | 12 | Disminuye | GTA | AX94983266, AX94480950, AX94590370 | ** | ns | * | * | ns | * | * | ns | * | ** | * | _ | Basile et al., 2019 |
| DE | B59-Hap3 | 3A | 695,5 | 695,7 | 19 | Aumenta | GA | AX94460524, AX94829707 | ns | * | ns | ns | ns | ** | * | ** | * | * | ns | _ | Basile et al., 2019 |
| DE | B60-Hap4 | 3A | 699,4 | 700,6 | 10 | Aumenta | GACG | AX95083017, AX94700391, AX94586183, AX95230073 | ns | ** | ns | ns | ns | ** | * | *** | ** | * | ns | _ | Basile et al., 2019 |
| DE | B7-Hap1 | 3D | 518,2 | 560,1 | 54 | Disminuye | CGCGAATATCTAGCCT | AX94918512, AX94838180, AX95014681, AX94749865, AX94643137, AX94629154, AX94462026, AX94508902, AX94762786, AX94401224, AX94804577, AX94505336, AX94524892, AX95078497, AX94475658, AX94965337 | * | * | ns | ns | ns | * | * | ** | * | * | * | _ | Basile et al., 2019 |
| DE | B75-Hap4 | 5B | 597,2 | 601,4 | 11 | Disminuye | TATGTTCGCTTCCG | AX94472448, AX94825873, AX95126299, AX94494539, AX94534515, AX94435670, AX94419463, AX94526460, AX94855957, AX95234715, AX95141741, AX95257158, AX94955787, AX95131986 | * | ns | ns | ns | ns | * | * | ns | * | ** | ns | _ | Basile et al., 2019 |
| DE | AX94610041 | 6A | 13,8 | - | 46 | _ | A | AX94610041 | * | * | ** | * | * | ** | ns | * | * | * | ns | _ | Basile et al., 2019 |
| DE | B43-Hap1 | 6A | 591,7 | 592,5 | 61 | Disminuye | TGTACT | AX94439933, AX94621070, AX95152390, AX94499400, AX94765421, AX94750793 | ns | ** | ns | ns | ns | * | * | ** | * | * | * | _ | Basile et al., 2019 |
| DE | B26-Hap3 | 6B | 157,6 | 157,8 | 11 | Aumenta | ACG | AX95231652, AX94976370, AX94918690 | * | ** | ns | ns | ns | ns | ns | ns | * | * | ** | PH | Basile et al., 2019 |
| DE | B24-Hap2 | 7A | 68 | 70,2 | 8 | Aumenta | CAAC | AX94880471, AX94952518, AX94923482, AX94915156 | ns | ns | * | ** | * | * | ns | ** | * | * | ns | _ | Basile et al., 2019 |
| AP | B38-Hap3 | 1A | 508,6 | 510,4 | 16 | Aumenta | CTAATTTG | AX95194961, AX94775918, AX94519549, Glu-A1_2*, Glu-A1_1, Glu-A1_null, AX94509092, AX95192108 | * | ** | ns | ns | ns | * | ns | ns | * | ns | ns | _ | Basile et al., 2019 |
| AP | B17-Hap4 | 1B | 121 | 124 | 5 | Aumenta | C-TAACAC | AX94479105, AX94923277, AX95215734, AX94819793, AX95630584, AX95233201, AX95117274, AX94511623 | * | * | * | ** | * | *** | * | * | * | * | ** | _ | Basile et al., 2019 |
| AP | AX94510167 | 1B | 548,7 | - | 21 | Aumenta | C | AX94510167 | ns | * | ns | ** | * | ** | ns | ns | * | ns | ns | _ | Basile et al., 2019 |
| AP | B54-Hap1 | 1B | 580,5 | 581,2 | 43 | Disminuye | GAT | AX94417290, AX94963680, AX94858556 | * | ** | ns | * | * | * | ns | ns | *** | *** | ns | _ | Basile et al., 2019 |
| AP | Lr10 | 1D | 8,6 | - | 10 | Aumenta | TTA | Lr 10_362 | * | * | ** | ns | * | ns | * | ns | * | * | ns | _ | Basile et al., 2019 |
| AP | B5-Hap4 | 1D | 11,6 | 11,9 | 19 | Disminuye | TCG | AX95069958, AX94514240, AX95653181 | * | ** | ns | * | * | * | ns | * | * | * | * | _ | Basile et al., 2019 |
| AP | B10-Hap4 | 1D | 31,1 | 32,5 | 10 | Aumenta | TTGC | AX94450337, AX94886765, AX94603350, AX95235028 | * | ns | *** | * | ns | * | * | ns | ns | ns | ns | _ | Basile et al., 2019 |
| AP | B39-Hap1 | 2A | 611,2 | 611,9 | 70 | Disminuye | ACCG | AX94929514, AX95166962, AX95076860, AX95238815 | ** | ns | ** | ns | ns | * | ns | * | ns | ns | * | _ | Basile et al., 2019 |
| AP | B48-Hap3 | 2A | 701 | 702,1 | 15 | Disminuye | TGAAT | AX94881363, AX94401392, AX94619928, AX94526179, AX94486441 | * | *** | * | ns | * | * | ns | ns | * | * | ns | _ | Basile et al., 2019 |
| AP | B56-Hap4 | 2B | 469,2 | 476,6 | 6 | Aumenta | C- | AX94917137, AX95133917 | ns | * | ** | * | ns | * | ns | * | ns | ** | * | _ | Basile et al., 2019 |
| AP | B79-Hap3 | 2B | 664,2 | 664,9 | 5 | Aumenta | TA- | AX94950749, AX95199286, AX95069755 | * | ns | ** | * | ns | ** | ns | * | ns | ** | * | _ | Basile et al., 2019 |
| AP | AX94687989 | 2B | 702,2 | - | 52 | Aumenta | T | AX94687989 | ns | ** | ns | ns | ns | ns | * | ns | ** | * | ns | _ | Basile et al., 2019 |
| AP | B97-Hap1 | 2B | 719,5 | 719,5 | 38 | Disminuye | CAT | AX94645156, AX94982580, AX94706037 | ns | ** | ns | ns | ns | ns | * | ns | ** | * | * | _ | Basile et al., 2019 |
| AP | B114-Hap4 | 2B | 762 | 763,9 | 8 | Aumenta | C-AAA | AX94868242, AX94431230, AX95097760, AX95191464, AX94912106 | ** | * | * | ns | * | ns | ns | ns | * | ns | ns | _ | Basile et al., 2019 |
| AP | AX94585596 | 2B | 776,4 | - | 15 | Aumenta | C | AX94585596 | * | ** | ns | ns | * | ** | * | * | * | ns | ns | _ | Basile et al., 2019 |
| AP | AX94624485 | 2B | 776,8 | - | 10 | Aumenta | G | AX94624485 | * | * | * | ns | ns | ** | * | * | ns | ns | ns | _ | Basile et al., 2019 |
| AP | B122-Hap3 | 2B | 777,1 | 777,2 | 6 | Aumenta | -A | AX94869081, AX94764862 | * | * | * | ns | * | *** | ** | * | * | * | ns | _ | Basile et al., 2019 |
| AP | B22-Hap2 | 3A | 391,5 | 393,4 | 53 | Aumenta | GC | AX94393045, AX94823297 | * | * | ns | * | ns | * | ** | * | * | ns | * | _ | Basile et al., 2019 |
| AP | AX95090222 | 3A | 700,6 | - | 18 | Aumenta | G | AX95090222 | ** | ns | ns | * | ns | ns | ** | ns | * | ns | ns | _ | Basile et al., 2019 |
| AP | AX95083205 | 3A | 743,9 | - | 46 | Aumenta | C | AX95083205 | ** | * | ns | ns | * | ns | ns | * | * | ns | ns | _ | Basile et al., 2019 |
| AP | B6-Hap1 | 3B | 164,4 | 170,8 | 65 | Disminuye | TCACG | AX94415818, AX94512569, AX94586240, AX94707486, AX95629899 | * | * | * | ns | * | ns | * | * | ** | ** | * | _ | Basile et al., 2019 |
| AP | B55-Hap4 | 3B | 511,1 | 513,6 | 6 | Disminuye | CCCTT | AX94541525, AX94453315, AX94990396, AX94853100, AX94547219 | ** | * | ns | ns | * | ns | ns | ns | ns | * | * | _ | Basile et al., 2019 |
| AP | B60-Hap6 | 3B | 541,5 | 544,5 | 5 | Aumenta | GA-G-CC-T | AX95141130, AX95685009, AX94701179, AX95630480, AX94829564, AX94900436, AX94434519, AX94445502, AX94929428 | * | ns | * | * | ns | ** | * | * | * | * | ns | _ | Basile et al., 2019 |
| AP | B109-Hap2 | 3B | 796 | 799,1 | 9 | Disminuye | CTGTTGACGATG | AX95681901, AX94724702, AX95085345, AX94613383, AX95229979, AX94460257, AX95630742, AX94991264, AX94967827, AX95210310, AX94528506, AX94512236 | * | ** | * | ns | ns | ns | ns | ns | ns | * | * | _ | Basile et al., 2019 |
| AP | B111-Hap4 | 3B | 817,4 | 817,8 | 11 | Aumenta | CCCC | AX94758693, AX94799334, AX94401378, AX95131891 | *** | ns | * | * | ** | *** | * | ** | * | * | * | TKW | Basile et al., 2019 |
| AP | B57-Hap6 | 4A | 738,7 | 739,7 | 9 | Disminuye | ATTAG | AX94416122, AX95249633, AX95150738, AX95652066, AX94511022 | * | ns | ns | ** | ns | ** | ns | ns | ns | * | ns | _ | Basile et al., 2019 |
| AP | B28-Hap5 | 4B | 612,2 | 613,2 | 6 | Aumenta | GG-G | AX94617210, AX94652395, AX94783372, AX94750774 | * | ** | * | * | * | * | * | * | * | * | ns | _ | Basile et al., 2019 |
| AP | B36-Hap2 | 4B | 650,7 | 650,7 | 57 | Disminuye | TGT | AX94484839, AX95096924, AX94521391 | ** | * | * | * | ns | * | ** | ** | ns | * | ns | _ | Basile et al., 2019 |
| AP | B16-Hap4 | 5A | 33 | 33,2 | 5 | Aumenta | CCC | AX94653931, AX94556600, AX94635713 | * | * | * | * | * | ** | ** | ns | ns | ns | ns | _ | Basile et al., 2019 |
| AP | B33-Hap4 | 5A | 445,2 | 445,2 | 7 | Disminuye | GC | AX95632187, AX94682497 | ns | *** | * | ns | ns | * | ns | ns | ns | * | * | FE | Basile et al., 2019 |
| AP | B54-Hap3 | 5A | 516,1 | 527,9 | 5 | Aumenta | CACTGAAG | AX94397871, AX94509108, AX94908260, AX94415346, AX94606396, AX94462177, AX95629459, AX94651425 | ns | * | * | ** | ** | *** | ns | ns | * | * | * | _ | Basile et al., 2019 |
| AP | B82-Hap2 | 5A | 611,6 | 613,5 | 10 | Aumenta | CACC | AX94413482, AX94423469, AX94437035, AX94747224 | * | ns | * | ns | ns | * | *** | ns | * | ns | * | _ | Basile et al., 2019 |
| AP | B26-Hap2 | 5B | 430,7 | 430,7 | 17 | Aumenta | CG | AX94432329, AX94784692 | ** | * | ns | * | * | * | * | ns | ns | ns | * | _ | Basile et al., 2019 |
| AP | AX94550178 | 5B | 455,7 | - | 46 | Aumenta | G | AX94550178 | ns | ns | *** | * | * | * | ns | ns | * | ns | ns | _ | Basile et al., 2019 |
| AP | B92-Hap4 | 5B | 683,5 | 684 | 5 | Aumenta | C- | AX94861020, AX94597114 | * | ns | * | * | ns | ** | * | * | * | ns | ** | _ | Basile et al., 2019 |
| AP | B2-Hap5 | 6A | 0,6 | 0,8 | 5 | Aumenta | -C | AX95630434, AX94746756 | * | ns | ** | ** | * | ns | ns | ns | * | * | ns | _ | Basile et al., 2019 |
| AP | B4-Hap2 | 6A | 2,2 | 2,7 | 26 | Aumenta | CGCC | AX94786709, AX95630503, AX95209203, AX95148675 | * | ns | * | * | ** | ns | * | ns | ns | * | ns | _ | Basile et al., 2019 |
| AP | B4-Hap3 | 6A | 2,2 | 2,7 | 17 | Disminuye | CATC | AX94786709, AX95630503, AX95209203, AX95148675 | ** | * | * | ns | * | ns | * | ns | * | ** | * | _ | Basile et al., 2019 |
| AP | B16-Hap1 | 6A | 48,6 | 49,8 | 57 | Aumenta | CCC | AX94889521, AX95233516, AX94739464 | ns | ns | ns | * | ns | * | * | ns | ** | * | ** | _ | Basile et al., 2019 |
| AP | B17-Hap3 | 6A | 50,3 | 51,4 | 33 | Disminuye | CCA | AX94472732, AX95184774, AX94579590 | ns | * | * | * | ns | ** | ns | * | * | ** | ns | _ | Basile et al., 2019 |
| AP | B54-Hap5 | 6A | 610 | 610,2 | 6 | Aumenta | -G | AX94900530, AX95188728, AX94554119 | ns | ns | * | * | ns | *** | ns | ** | ns | ns | ns | _ | Basile et al., 2019 |
| AP | B7-Hap2 | 6B | 19,8 | 20,7 | 82 | Disminuye | CAC | AX94606720, AX95152971, AX94679404 | ** | *** | ** | * | ns | * | *** | * | * | * | * | _ | Basile et al., 2019 |
| AP | B20-Hap4 | 6B | 124,4 | 126,1 | 6 | Aumenta | CCC | AX94725996, AX95154560, AX95069655 | * | ns | ns | ns | ns | ** | * | * | ns | * | * | _ | Basile et al., 2019 |
| AP | B25-Hap4 | 6B | 151,4 | 156,4 | 25 | Disminuye | TCATAGGCCTTGAGTTC | AX94476270, AX94489861, AX94594212, AX94592155, AX95228304, AX94746774, AX95251482, AX95224228, AX94436738, AX94925666, AX94723474, AX95257540, AX95254919, AX94814333, AX95150132, AX94933359, AX94606831 | * | ** | * | * | ** | ns | ns | ns | * | * | ns | _ | Basile et al., 2019 |
| AP | B26-Hap3 | 6B | 157,6 | 157,8 | 11 | Disminuye | ACG | AX95231652, AX94976370, AX94918690 | * | *** | ns | * | ns | * | * | * | ns | * | *** | HD | Basile et al., 2019 |
| AP | B31-Hap2 | 6B | 195,3 | 197,8 | 5 | Aumenta | -G | AX94507593, AX94510067 | * | ns | ** | ** | * | ** | ns | ns | * | * | * | _ | Basile et al., 2019 |
| AP | AX94943227 | 6B | 234,8 | - | 17 | Aumenta | C | AX94943227 | ** | *** | ** | * | * | * | *** | * | * | * | * | _ | Basile et al., 2019 |
| AP | B47-Hap3 | 6B | 480,4 | 491,6 | 24 | Disminuye | GGTCGTTGCCAGC | AX94485266, AX94999452, AX94504202, AX94501866, AX94417160, AX94459292, AX94596473, AX94878625, AX94647376, AX94523508, AX94690961, AX94916954, AX95197645 | ns | ns | ns | * | ** | ns | ns | ns | * | ns | * | _ | Basile et al., 2019 |
| AP | AX94476474 | 7A | 561,6 | - | 14 | Aumenta | T | AX94476474 | ** | * | * | ns | ns | ns | ns | ** | * | * | ns | _ | Basile et al., 2019 |
| AP | B14-Hap4 | 7B | 59,6 | 61,6 | 22 | Disminuye | ATAC | AX94590671, AX94587603, AX94758128, AX95222995 | *** | ns | ns | ns | * | ns | * | ns | ns | * | ns | _ | Basile et al., 2019 |
| AP | B76-Hap3 | 7B | 743,5 | 743,6 | 10 | Disminuye | CA | AX94593529, AX94770939 | ** | *** | * | * | ns | * | ns | * | ** | ** | * | _ | Basile et al., 2019 |
En la tabla solo se presentan los SNP que pasaron los criterios de selección en GWAS. ns = no significativo, * P <0.05, ** P <0.001, *** P <0.0001, “–” = no disponible.
Referencias
- Barrett, J. C., Fry, B., Maller, J., & Daly, M. J. (2004). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21(2), 263–265. https://doi.org/10.1093/bioinformatics/bth457
- Basile, S. M. L., Ramírez, I. A., Crescente, J. M., Conde, M. B., Demichelis, M., Abbate, P., Rogers, W. J., Pontaroli, A. C., Helguera, M., & Vanzetti, L. S. (2019). Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biology, 19(1). https://doi.org/10.1186/s12870-019-2015-4
- François O. (2016). Running structure-like population genetic analyses with R. http://membres-timc.imag.fr/Olivier.Francois/tutoRstructure.pdf
- Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., Gore, M. A., Buckler, E. S., & Zhang, Z. (2012). GAPIT: genome association and prediction integrated tool. Bioinformatics, 28(18), 2397–2399. https://doi.org/10.1093/bioinformatics/bts444
- Vanzetti, L. S., Yerkovich, N., Chialvo, E., Lombardo, L., Vaschetto, L., & Helguera, M. (2013). Genetic structure of Argentinean hexaploid wheat germplasm. Genetics and Molecular Biology, 36(3), 391–399. https://doi.org/10.1590/s1415-47572013000300014
- Wang, Q., Tian, F., Pan, Y., Buckler, E. S., & Zhang, Z. (2014). A SUPER Powerful Method for Genome Wide Association Study. PLoS ONE, 9(9), e107684. https://doi.org/10.1371/journal.pone.0107684
- Zhang, J., Gizaw, S. A., Bossolini, E., Hegarty, J., Howell, T., Carter, A. H., Akhunov, E., & Dubcovsky, J. (2018). Identification and validation of QTL for grain yield and plant water status under contrasting water treatments in fall-sown spring wheats. Theoretical and Applied Genetics, 131(8), 1741–1759. https://doi.org/10.1007/s00122-018-3111-9